CASE STUDIES
Transforming our ability to understand and manage our environment.
Examples of the Genomics work supported by NEOF and the Liverpool and Sheffield nodes of the NERC Biomolecular Analysis Facility (the predecessor to NEOF) from 2019.
Examples of Proteomics work supported by the Centre for Proteomics Research, University of Liverpool.
Examples of Metabolomics work supported by the Centre for Proteomics Research, University of Liverpool.

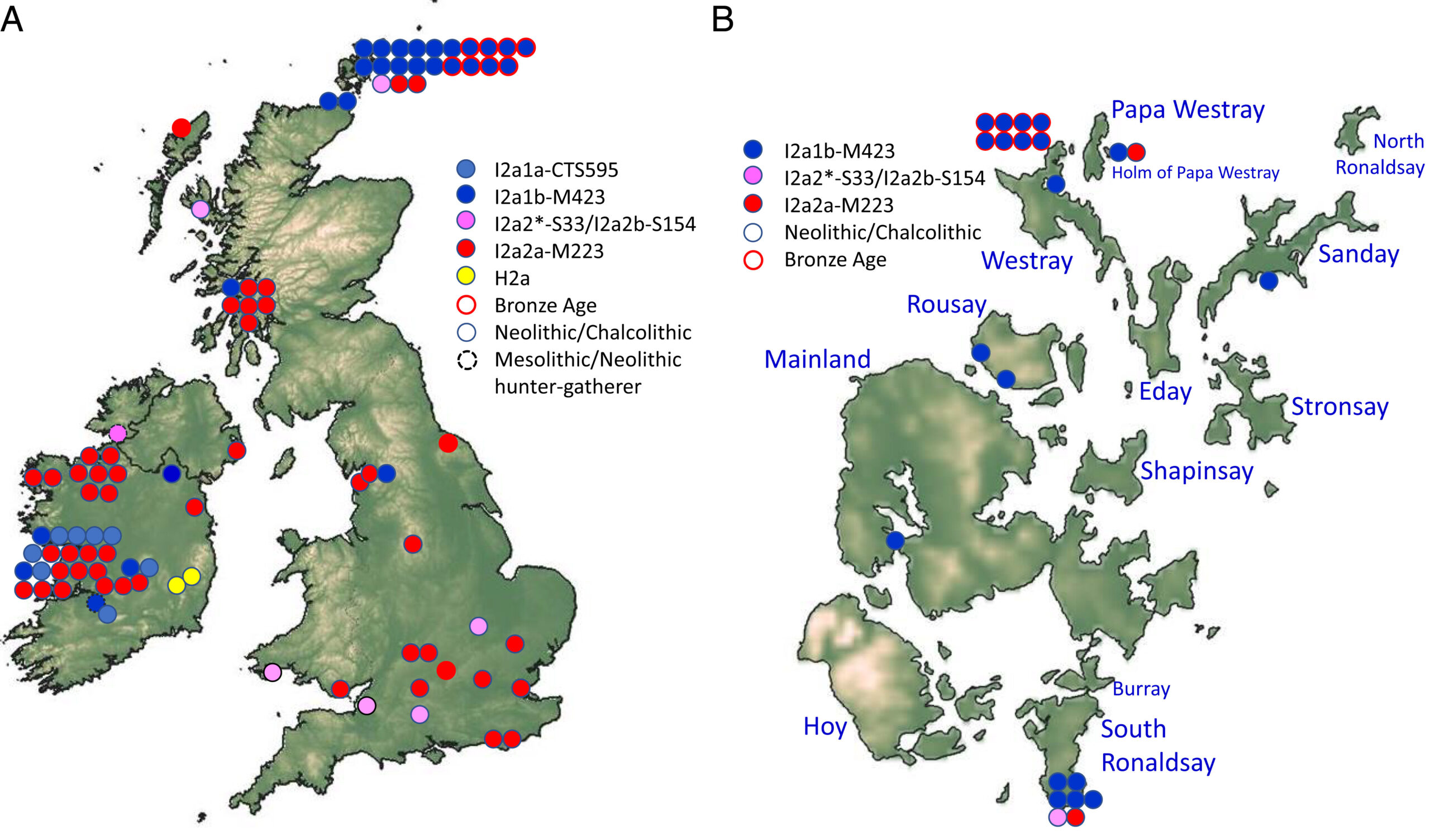
Ancient DNA at the edge of the world: Continental immigration and the persistence of Neolithic male lineages in Bronze Age Orkney (2022)
Genome sequencing by NEOF helped to show that Orkney in the Bronze Age showed far more immigration, and hence movement of both people and culture, than previously believed. It was particularly striking that immigration was almost solely of females, with male Y lineages persisting for at least a thousand years.
Dulias et al (2022) PNAS 10.1073/pnas.2108001119
Invisible
READ MORE
Orkney was a major cultural center during the Neolithic, 3800 to 2500 BC. Farming flourished, permanent stone settlements and chambered tombs were constructed, and long-range contacts were sustained. From ∼3200 BC, the number, density, and extravagance of settlements increased, and new ceremonial monuments and ceramic styles, possibly originating in Orkney, spread across Britain and Ireland. By ∼2800 BC, this phenomenon was waning, although Neolithic traditions persisted to at least 2500 BC. Unlike elsewhere in Britain, there is little material evidence to suggest a Beaker presence, suggesting that Orkney may have developed along an insular trajectory during the second millennium BC. We tested this by comparing new genomic evidence from 22 Bronze Age and 3 Iron Age burials in northwest Orkney with Neolithic burials from across the archipelago. We identified signals of inward migration on a scale unsuspected from the archaeological record: As elsewhere in Bronze Age Britain, much of the population displayed significant genome-wide ancestry deriving ultimately from the Pontic-Caspian Steppe. However, uniquely in northern and central Europe, most of the male lineages were inherited from the local Neolithic. This suggests that some male descendants of Neolithic Orkney may have remained distinct well into the Bronze Age, although there are signs that this had dwindled by the Iron Age. Furthermore, although the majority of mitochondrial DNA lineages evidently arrived afresh with the Bronze Age, we also find evidence for continuity in the female line of descent from Mesolithic Britain into the Bronze Age and even to the present day.
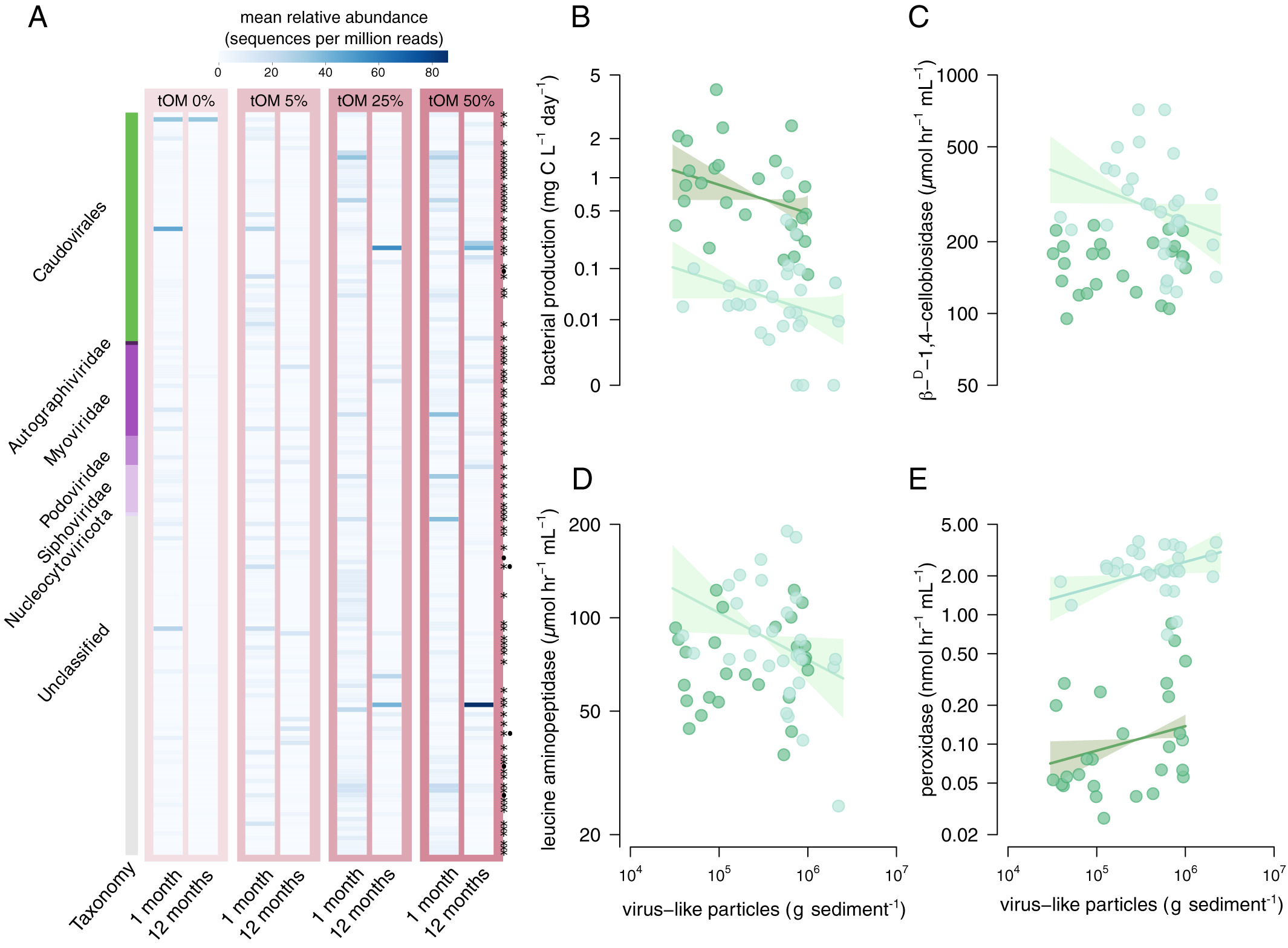
Viruses direct carbon cycling in lake sediments under global change (2022)
eDNA analysis at NEOF helped to show the importance of viruses to carbon cycling in lake sediments and to identify viral genes that affect the processing of carbon compounds. These results argue that viruses need to be included in any predictive model of the freshwater carbon cycle in response to climate change.
(Braga et al 2022 PNAS)
Invisible
READ MORE
Global change is altering the vast amount of carbon cycled by microbes between land and freshwater, but how viruses mediate this process is poorly understood. Here, we show that viruses direct carbon cycling in lake sediments, and these impacts intensify with future changes in water clarity and terrestrial organic matter (tOM) inputs. Using experimental tOM gradients within sediments of a clear and a dark boreal lake, we identified 156 viral operational taxonomic units (vOTUs), of which 21% strongly increased with abundances of key bacteria and archaea, identified via metagenome-assembled genomes (MAGs). MAGs included the most abundant prokaryotes, which were themselves associated with dissolved organic matter (DOM) composition and greenhouse gas (GHG) concentrations. Increased abundances of virus-like particles were separately associated with reduced bacterial metabolism and with shifts in DOM toward amino sugars, likely released by cell lysis rather than higher molecular mass compounds accumulating from reduced tOM degradation. An additional 9.6% of vOTUs harbored auxiliary metabolic genes associated with DOM and GHGs. Taken together, these different effects on host dynamics and metabolism can explain why abundances of vOTUs rather than MAGs were better overall predictors of carbon cycling. Future increases in tOM quantity, but not quality, will change viral composition and function with consequences for DOM pools. Given their importance, viruses must now be explicitly considered in efforts to understand and predict the freshwater carbon cycle and its future under global environmental change.
Braga et al (2022) Viruses direct carbon cycling in lake sediments under global change. PNAS.
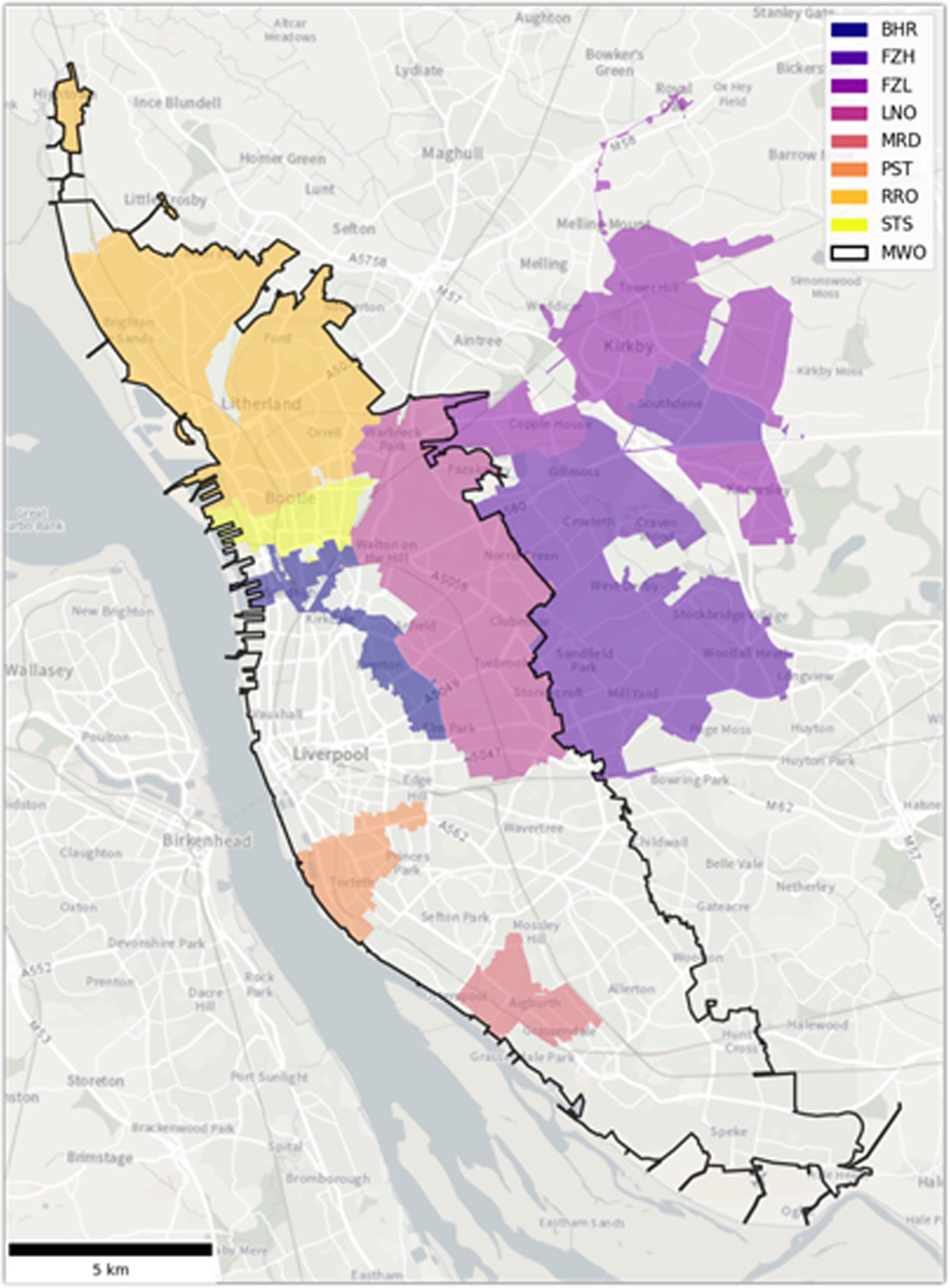
City-wide wastewater genomic surveillance through the successive emergence of SARS-CoV-2 Alpha and Delta variants (2022)
NEOF expertise and capability was used to develop and implement wastewater based epidemiology to detect and monitor SARS-CoV-2 variants. Here we demonstrated this capability to detect the emergence and rise of Alpha and Delta variants within an urban sewage system that matched clinical sampling but at a fraction of the cost.
Brunner et al (2022) Water Research 226, 119306
Invisible
READ MORE
Genomic surveillance of SARS-CoV-2 has provided a critical evidence base for public health decisions throughout the pandemic. Sequencing data from clinical cases has helped to understand disease transmission and the spread of novel variants. Genomic wastewater surveillance can offer important, complementary information by providing frequency estimates of all variants circulating in a population without sampling biases. Here we show that genomic SARS-CoV-2 wastewater surveillance can detect fine-scale differences within urban centres, specifically within the city of Liverpool, UK, during the emergence of Alpha and Delta variants between November 2020 and June 2021. Furthermore, wastewater and clinical sequencing match well in the estimated timing of new variant rises and the first detection of a new variant in a given area may occur in either clinical or wastewater samples. The study’s main limitation was sample quality when infection prevalence was low in spring 2021, resulting in a lower resolution of the rise of the Delta variant compared to the rise of the Alpha variant in the previous winter. The correspondence between wastewater and clinical variant frequencies demonstrates the reliability of wastewater surveillance. However, discrepancies in the first detection of the Alpha variant between the two approaches highlight that wastewater monitoring can also capture missing information, possibly resulting from asymptomatic cases or communities less engaged with testing programmes, as found by a simultaneous surge testing effort across the city.
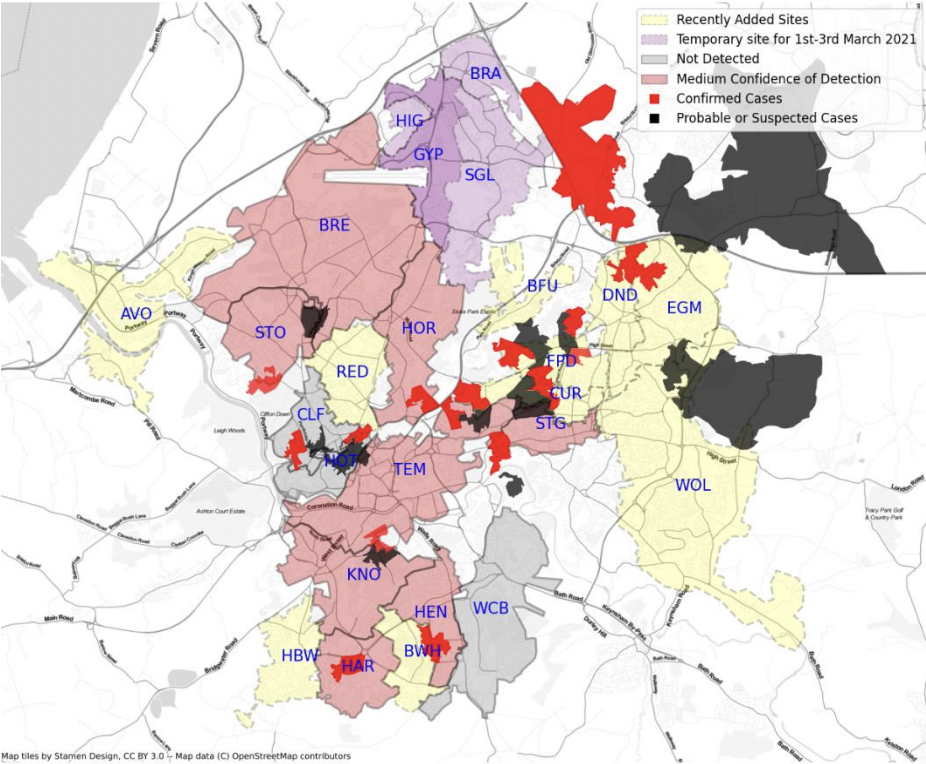
Environmental monitoring of SARS-CoV-2 (2021)
NEOF led the design of sequencing assays and generation data and analysis.
- Hilary et al (2021) Water Research 200, 117214
- Brown et al (2021) Summary for SAGE
Invisible
READ MORE
In support of the UK national response to COVID-19, NEOF prioritised work to use our skills in eDNA to detect and characterise coronavirus in wastewater. Working with the University of Bangor, we were able to demonstrate that SARS-CoV-2 could be genome-sequenced from wastewater taken from treatment plants, and were able to characterise its genetic diversity through the first wave of the pandemic. Having demonstrated the potential of the method, we then worked with UK Health Security Agency (UKHSA) Monitoring Health Protection Programme to deliver national surveillance of variants in wastewater for more than 1000 samples per week. This initial work by NEOF led to a major consortium of sequencing laboratories supported by funding from UKHSA. Highlights have included a rapid report to the CMO of the alpha (“Kent”) variant present in samples from London through November and December 2020, and an outbreak of an E484K mutation in Bristol used as a case study in a paper to SAGE.

Bumblebee colony density on farmland (2021)
NEOF supported training in microsatellite genotyping and data analysis at the visitor facility. (Timberlake et al, 2021 JAE 1016-16)
Invisible
READ MORE
NEOF provided support and training to a PhD student project in the NEOF Visitor Facility to understand how the landscape affects the density of bumblebees, which are an important pollinator providing an essential ecosystem function. Molecular genotyping was used to estimate colony density on farmland and how this varied with nectar supply and garden cover. These results discovered the importance of the late spring as a bottleneck for resources, and highlighted the role that environmental stewardship programmes of farmland could have in increasing the density of bumblebee colonies, for example by modifying mowing regimes to delay flowering at field edges. This provides an example of how omics methods can provide an environmental solution within a systems approach. This work involved both international partners and conservation bodies, and received media interest, including in the farming press.
Timberlake et al. (2021) Bumblebee colony density on farmland is influenced by late-summer nectar supply and garden coverJ Appl Ecol 58, 1006–1016.

Gradients in richness and turnover of a forest passerine’s diet prior to breeding: A mixed model approach applied to faecal metabarcoding data (2020)
NBAF supported metabarcoding and bioinformatic analysis.
Invisible
READ MORE

Meiotic drive reduces egg-to-adult viability in stalk-eyed flies (2019)
NBAF supported microsatellite genotyping and data analysis.
Invisible
READ MORE
Several species exhibit the phenomenon of Sex-Ratio (SR) meiotic drive, a selfish genetic element located on the X-chromosome that causes dysfunction of Y-bearing sperm, and one that has the potential to be exploited in the control of harmful insect vectors such as some mosquitoes. SR is transmitted to up to 100% of offspring, causing extreme sex-ratio bias. SR in several species is found in a stable polymorphism, suggesting that there must be strong frequency-dependent selection resisting its spread. This study investigated the effect of SR on female and male egg-to-adult viability in the Malaysian stalk-eyed fly, Teleopsis dalmanni. SR meiotic drive in this species is old, and appears to be broadly stable at a moderate (ca 20%) frequency. Large-scale controlled crosses were used to estimate the strength of selection acting against SR in female and male carriers. SR was found to reduce the egg-to-adult viability of both sexes. In females, homozygous females experienced greater reduction in viability and the deleterious effects of SR were additive. The male deficit in viability was not different from that in homozygous females. The study contributed to our understanding of how these reductions in egg-to-adult survival maintain the SR polymorphism in this species.

Individual variation in early-life telomere length and survival in a wild mammal (2019)
NBAF supported telomere length analysis using quantitative PCR methods.
Invisible
READ MORE

Parallel evolution of complex centipede venoms revealed by comparative proteotranscriptomic analyses (2019)
NBAF supported transcriptome sequencing.
Invisible
READ MORE

Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems (2019)
NBAF supported metabarcoding.
Invisible
READ MORE

Lack of long-term acclimation in Antarctic encrusting species suggests vulnerability to warming (2019)
NBAF supported RNA-seq and metabarcoding.
Invisible
READ MORE
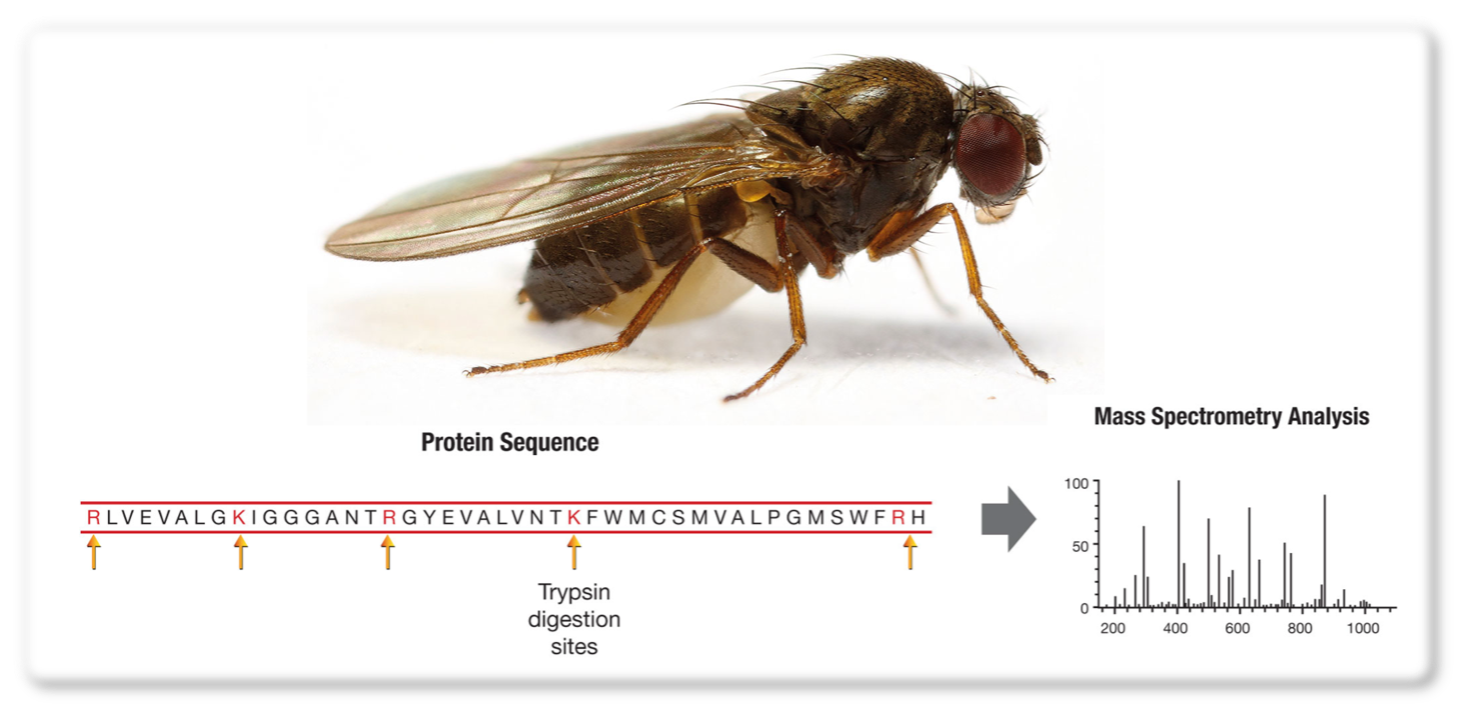
Proteomics case study: Targeting specific proteins or peptides
Aim: To identify sex peptides from Drosophila subobscura accessory gland samples.
Samples: 3 x 10 excised excretory glands from D. subobscura.
Results: There is no protein database for D. sub in UniProt, However, we were able to identify a suitable database by contacting other researchers who work with this organism. In addition, by pre-calculating the expected masses of trypsin cleavage products from the specific sex peptides of interest, we were able to perform target-ed analysis of these peptides to get a positive identification.
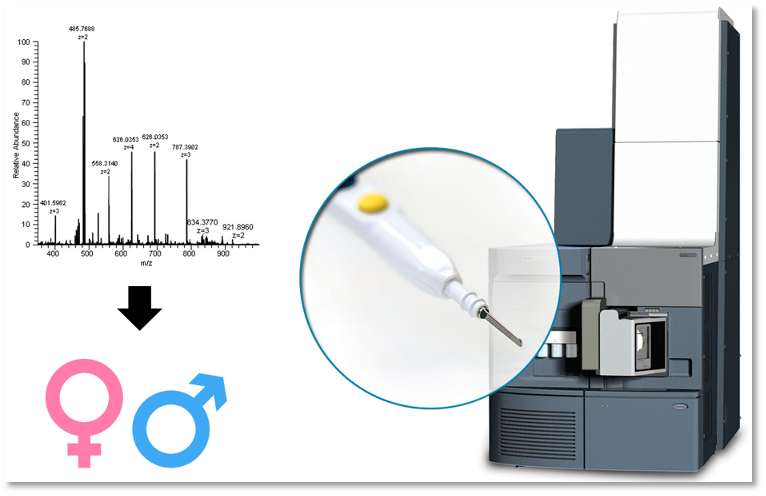
Proteomics case study: Species identification
Aim: Rapid identification of species and sex, based on pattern recognition of mass spectra, for closely related species that are notoriously difficult to identify by eye.
Samples: 150-170 whole mosquito samples of each of D. bifasciata, D. pseudoobscura, D. suboscura, D. melanogaster and D. simulans were used to build a species model; 400 male and 400 female mosquitoes across the 5 species were used to build the sex determination model.
Results: 95 % accuracy in species determination, and 99 % accuracy in sex determination.
Proteomics case study: Impact of viral infection on host proteome
Aim: To quantify changes to the Asian tiger mosquito proteome when exposed to virus.
Samples: 3 replicates of 14 conditions.
Results: By using TMT isobaric labelling, more than 5000 proteins were quantified including 11 out of 12 virus proteins. Proteins were identified from a genome database.
Proteomics case study: Secretome proteome of pseudomonas aeruginosa
Aim: To identify the difference in multi-drug resistance strain verse the wild type.
Samples: 5 replicates of 2 pseudomonas aeruginosa strains.
Results: Proteins were concentrated from 10 mL to 100 μL by strataclean beads. Digestion was conducted from straight from the strataclean beads. Over 1000 proteins were identified in each sample. Samples were processed for label free quantification.
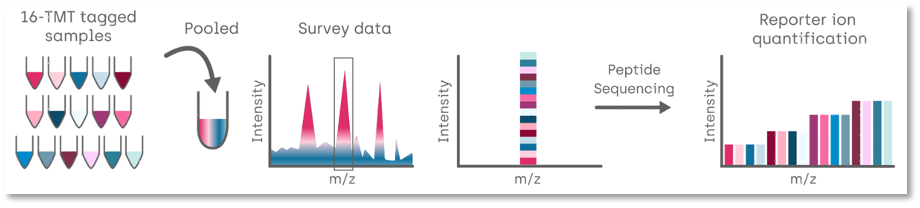
Proteomics case study: Identification of phosphorylation targets for a phosphatase in human cell line
Aim: To determine which proteins are phosphorylated and the down stream effects of phosphorylation.
Samples: 4 replicates of 4 clonal cell lines.
Results: Samples were labelled with TMT isobaric labelling. The sample was fractionated into 12 fractions and then each used to deter mine i) global protein changes and ii) phosphopeptide changes. Around 6000 proteins and in excess of 10,000 phosphopeptides were identified, with the vast majority being quantified across all 16 samples.

Metabolomics case study: probing the effects of climate change stress on Antarctic marine organisms
Biological interpretation by collaborators is ongoing, but this work offers insights into adaptations that favour protein synthesis and stability at cold temperatures, and data will support comparisons with non-polar marine animals to identify adaptation to low temperature. These experiments provide a proof of principle study, with the potential to answer fundamental questions about cellular level adaptation and homeostasis on marine species inhabiting the cryosphere that have so far been intractable.
Recipient of 2022 NEOF Independent Pilot Competition funding
Invisible
READ MORE
Small organic solutes (osmolytes) aid cells and tissues of marine organisms in balancing osmotic pressure and maintaining cell volume. Climate change in the polar regions has led to increased sea ice melt and falling salinity of localised areas of the Antarctic Ocean. The role of osmolytes and metabolic end products in long-term salinity stress tolerance of Sterechinus neumayeri and Odontaster validus was probed using an untargeted metabolomics experimental design approach.
The NEOF Centre for Metabolomics Research (CMR) directed experimental design and sample collection parameters to ensure robust data generation and achieve high metabolite yield. A series of optimisation experiments were carried out by the CMR lab to evaluate metabolite extraction, sample processing and instrument methods. A gas chromatography-mass spectrometry analysis approach was selected, based on the compounds of interest. Organisms were exposed to a native and two salinity stress conditions, with tissue and extracellular fluid collected at defined time points. The CMR’s versatile standard GC-MS method was used in analysis of the extracted samples, in accordance with Metabolomics Quality Assurance & Quality Control Consortium (mQACC) standards.
Mass spectrometry data processing and statistical analysis were also carried out by the CMR, including peak deconvolution, feature alignment, reference index assignment, spectral library based peak annotation and pooled QC/labelled internal standard based normalisation. Metabolome profiling revealed statistically significant fold changes in several key features within exposure groups, which indicate their use as osmolytes in the low salinity acclimation process. In addition, collaborators identified several novel compounds appearing to have cryoprotective and DNA protective properties.
Biological interpretation by collaborators is ongoing, but this work offers insights into adaptations that favour protein synthesis and stability at cold temperatures, and data will support comparisons with non-polar marine animals to identify adaptation to low temperature. These experiments provide a proof of principle study, with the potential to answer fundamental questions about cellular level adaptation and homeostasis on marine species inhabiting the cryosphere that have so far been intractable. The results of this pilot will aid application to wider grant funding for a large-scale project.
Collaborators Prof. Lloyd Peck and Nicholas Barrett (PhD Candidate) (British Antarctic Survey/University of Cambridge) were awarded pilot funding after a successful bid in the NEOF Independent Pilot Competition 2022. Data not yet published.
Metabolomics case study: Using metabolomics to understand how environmental bacteria respond to pharmaceutical exposure
Mol. BioSyst. 2016, 12, 1367. https://doi.org/10.1039/c5mb00889a
PLoS ONE 2016 11: e0156509. https://doi.org/10.1371/journal.pone.0156509
Invisible
READ MORE
It has been known for decades that human pharmaceuticals are present in wastewater treatment plants and pollute rivers and estuaries at levels that affect aquatic organisms (see PNAS paper). What isn’t know is how microbial communities respond to exposure to APIs (active pharmaceutical ingredients) and this is likely to be at the phenotypic level where the microorganism responds to this stress. In addition, bacteria and fungi may transform APIs into other substances that could also affect both the planktonic and benthic communities.
In a paper published in Molecular BioSystems we used a combination of metabolic fingerprinting using Fourier transform infrared (FT-IR) spectroscopy with metabolic profiling using gas chromatography-mass spectrometry (GC-MS) to investigate phenotypic changes in an environmental isolate of Pseudomonas putida. Initially this bacterium was exposed to six APIs (acetaminophen, atenolol, diclofenac, ibuprofen, mefenamic acid, propranolol) at levels that did not affect the growth profile of the organism. Focussing on propranolol (a β-blocker used to treat hypertension) we found significant changes in carbon metabolism and in particular alterations in the concentration of energy related metabolites, including ATP. In addition, we found profound changes in the saturation levels of cardiolipins which may be related to the synthesis and activation of solvent extrusion pumps which would eliminate propranolol from inside these bacterial cells.
We went onto conduct an in-depth metabolomics study focussing on the efflux pumps involved in solvent extrusion which was published in PLoS ONE. In this study we included a series of efflux pump knockout mutants of P. putida DOT-T1E to assess the effect of this transporter on propranolol and metabolism.
These studies have opened up the exciting prospect of understanding how microbial communities adapt when exposed to toxic materials.
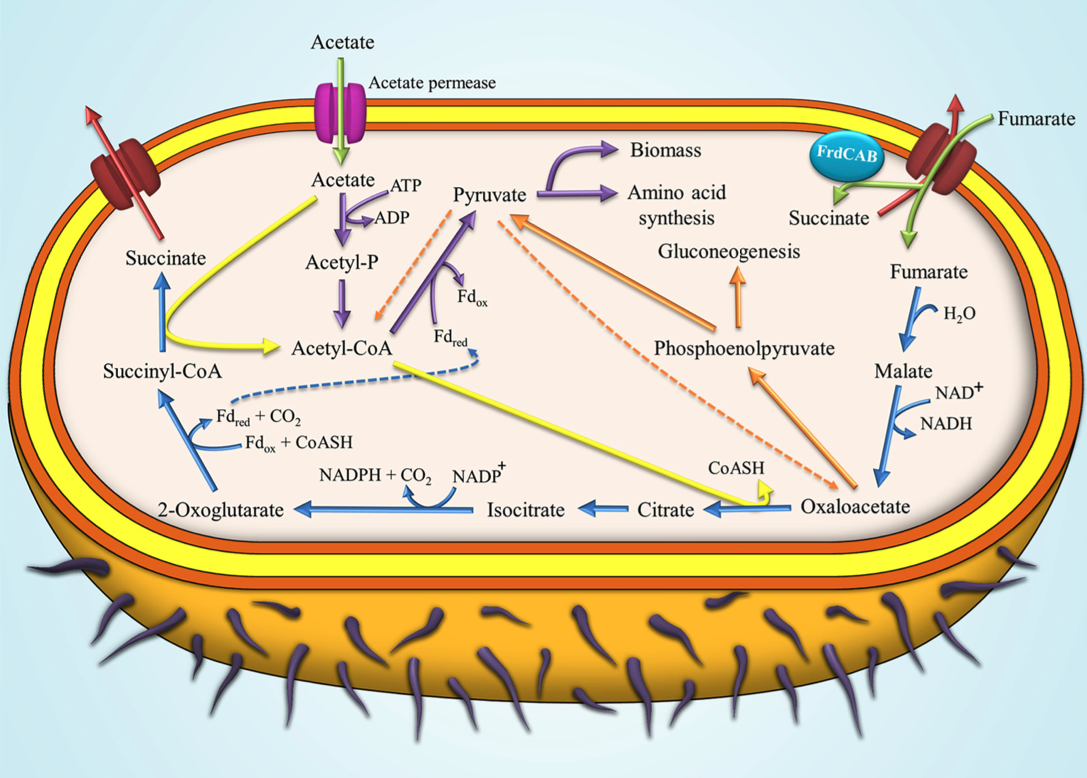
Metabolomics case study: Metabolic Profiling of Geobacter sulfurreducens during Industrial Bioprocess Scale-Up
Appl. Environ. Micro. 2015 81: 3288-3298. https://doi.org/10.1128/AEM.00294-15
Invisible
READ MORE
Geobacter sulfurreducens is an important bacterium as it is capable of global recycling of metals and therefore is important for bioremediation applications. In addition, this organism produces biogenic magnetite nanoparticles (BMNPs) which may have industrial application. However, during industrial scale-up of this BMNP production bioprocess we found an extended lag phase of 24 h for cells grown in 5-litre bioreactors compared to only 6 h those grown in 100 mL bottles.
In a paper published in Applied and Environmental Microbiology we investigated the phenotypic changes associated with the extended lag-phase during scale up of this bioprocess scale up using gas chromatography-mass spectrometry (GC-MS) for metabolic profiling.
GC-MS analysis showed that there were several changes in metabolites during scaleup and when relevant metabolites were overlaid onto a metabolic network of G. sulfurreducens we found that there was a limited availability of oxaloacetate and nicotinamide which would seem to be the main metabolic bottlenecks. Metabolite feeding experiments found that nicotinamide supplementation (1 mM) significantly reduced the lag phase in 5 L bioreactors from 24 h to 6 h, therefore resulting in a more rapid industrial bioprocess.
This study illustrates the exciting prospect of using metabolomics to enhance industrial scaleup of environmentally relevant microbial systems.
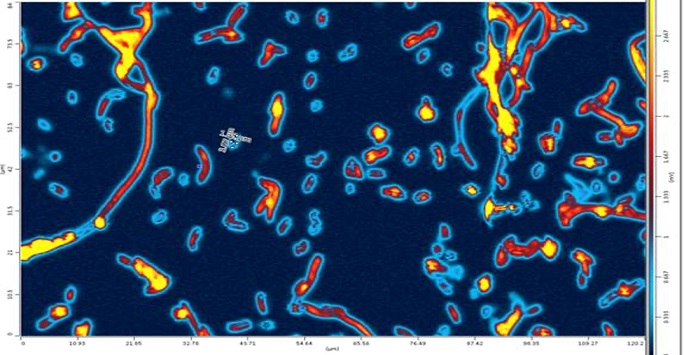
Metabolomics case study: Metabolism in Action: Developing stable isotope probing using Raman and infrared imaging for understanding metabolic flux at the single cell level
Anal. Chem. 2021, 93, 6, 3082–3088; https://doi.org/10.1021/acs.analchem.0c03967
Analyst 2021; 146, 1734-1746; https://doi.org/10.1039/D0AN02319A
Invisible
READ MORE
Complex microbial communities are found throughout ecosystems on the planet and these play essential functions that support animal and aquatic life.
Despite the vital role that bacteria play, linking these microbes with specific metabolic functions is difficult, since microbial communities consist of numerous and phylogenetically diverse microbes. This is compounded by the microscopic size of these microorganism (1-2 µm).
There is therefore a need to develop metabolomics solutions that generate images that are based on the biochemistry of the sample under analysis, and that provide information at micron and sub-micrometre scales. With the advent of spontaneous Raman and variants that can be tuned to specific vibrations (viz SRS and CARS), these, along with optical-photothermal infrared (O-PTIR) microscopy open up the tantalising opportunity to follow metabolism in microbial communities.

In a recent series of papers published in Analytical Chemistry and Analyst we have shown that the cellular uptake of stable isotope-labelled compounds by bacteria can be probed at the single-cell level using both Raman and infrared spectroscopies. This is due to the ability of these imaging techniques to monitor chemical vibrations that are affected by the incorporation of “heavy” atoms by cells and thus can be used to understand microbial systems. In this series of experiments, we have shown that it is possible to measure incorporation of 13C and 15N substrates, follow the dynamics of the rate of incorporation, as well as employing deuterated water to assess cross feeding in microbial communities.
This research has opened up the exciting prospect that these enabling chemical imaging technologies will guide the identification of primary substrate consumers in complex microbial communities in situ. This we believe represents a key step towards the characterisation of novel genes, enzymes and metabolic flux analysis in microbial consortia.
